Target base editing in soybean using a modified CRISPR/Cas9 system
作者:Yupeng Cai#, Li Chen#, Yan Zhang#, Shan Yuan#, Qiang Su, Shi Sun, Cunxiang Wu, Weiwei, Yao, Tianfu Han, Wensheng Hou*
影响因子:6.84
刊物名称:Plant Biotechnology Journal
出版年份:April 2020
doi: 10.1111/pbi.13386
https://onlinelibrary.wiley.com/doi/10.1111/pbi.13386
Abstract
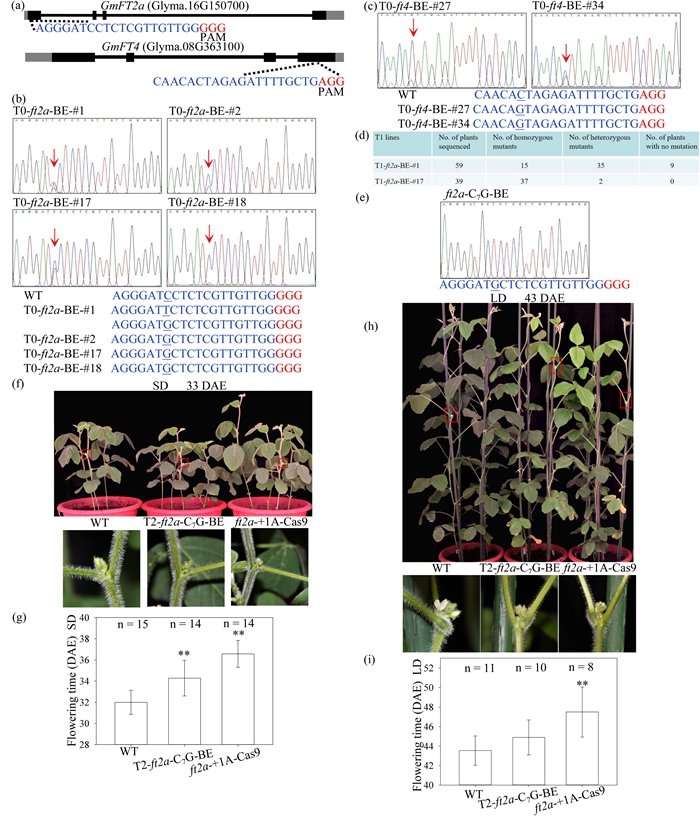
At present, the application of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat)/Cas9 (CRISPR-associated 9) system in soybean ( Glycine max ) has been mainly focused on knocking out target genes by frameshift mutations through non-homologous end joining. Many agriculturally important traits are associated with single nucleotide polymorphism (SNP) variation. Utilization of functional SNPs is also an important means to improve agronomic characters of crops. Therefore, generation of point mutations at specific sites associated with diverse important agronomic traits is of great value in molecular breeding. However, the recently developed base editing tool, which enables single base substitution into another, has not been systematically explored in soybean. In this study, we developed a CRISPR/Cas9-mediated base editing tool in soybean, and achieved specifically single base substitutions in target sites of two FLOWERING LOCUS T homologs, GmFT2a (Glyma.16G150700) and GmFT4 (Glyma.08G363100) with frequencies of 18.2% and 6.0% in the T0 transgenic plants, respectively. In addition, we demonstrated that these base editing events can be inherited. The ft2a mutants with a homozygous C to G change at target sequence exhibited later flowering phenotype, especially under short-day conditions. In summary, we have successfully introduced single base substitutions at target sites, indicating the feasibility of this base editing tool in soybean, which may expand the scope of application of CRISPR/Cas9 in soybean. Utilization of base editing technology holds great potential for future customized genetic improvement and breeding.